Self-aligning smart microscope achieves unprecedented resolution
Posted: 23 April 2020 | Hannah Balfour (Drug Target Review) | No comments yet
The developers of the ultra-precise single-molecule microscope demonstrated it can resolve interactions between molecules within living cells and is compatible with existing microscopes.

Researchers have developed a “smart single-molecule microscope” which can achieve unprecedented resolution capabilities and accurately detect interactions between individual molecules within intact cells.
The team from University of New South Wales (UNSW), Australia, worked off of research that won the 2014 Nobel Prize in Chemistry: the development of a super-resolution fluorescence microscopy technology that could view molecules inside living cells, enabling microscopists to explore complex biological systems and processes.
However, this microscopy technology was unable to observe interactions between molecules because its resolution capabilities were insufficient, with the interactions occurring at a scale at least four times smaller than that resolved by existing single-molecule microscopes.
“The reason why the localisation precision of single-molecule microscopes is around 20-30 nanometres normally is because the microscope actually moves while we are detecting that signal. This leads to an uncertainty. With the existing super-resolution instruments, we cannot tell whether or not one protein is bound to another protein because the distance between them is shorter than the uncertainty of their positions,” said Scientia Professor Katharina Gaus, research team leader and Head of UNSW Medicine’s EMBL Australia Node in Single Molecule Science.
To overcome the movement issue, the researchers built autonomous feedback loops inside a single-molecule microscope to detect and re-align the optical path and stage.
“It doesn’t matter what you do to this microscope, it basically finds its way back with precision under a nanometre. It’s a smart microscope. It does all the things that an operator or a service engineer needs to do and it does that 12 times per second,” explained Professor Gaus.
According to the team the feedback system is compatible with existing microscopes and allows for high flexibility in sample preparation. Professor Gaus said: “It’s a really simple and elegant solution to a major imaging problem. We just built a microscope within a microscope and all it does is align the main microscope. That the solution we found is simple and practical is a real strength as it would allow easy cloning of the system and rapid uptake of the new technology.”
Measuring the distance between proteins
Using their novel ultra-precise feedback single-molecule microscope, the team measured the distance between signalling proteins within T cells. It has been hypothesised by immunologists that the T cells are held in an inactive, or resting, state because signalling molecules within the cells interact with a receptor to prevent activation.
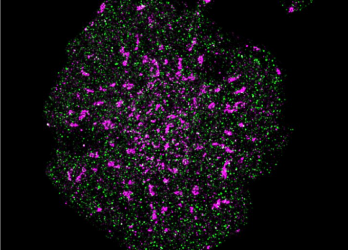

A T cell with precise localisation of T cell receptors (pink) and CD45 phosphatase (green) [credit: Single Molecule Science, UNSW Sydney].
Using this new technology, the team demonstrated that this is in fact the case, with the signalling molecule and T receptor being further separated from each other in activated T cells.
“Conventional microscopy techniques would not be able to accurately measure such a small change as the distance between these signalling molecules in resting T cells and in activated T cells only differed by 4-7 nanometres. This also shows how sensitive these signalling machineries are to spatial segregation. In order to identify regulatory processes like these, we need to perform precise distance measurements, and that is what this microscope enables. These results illustrate the potential of this technology for discoveries that could not be made by any other means,” concluded Professor Gaus.
The paper was published in Science Advances.
Related topics
Analytical Techniques, Disease Research, Drug Targets, Imaging, Microscopy, Molecular Biology, Molecular Targets, Technology
Related organisations
The University of New South Wales (UNSW)
Related people
Professor Katharina Gaus