Atomic-scale details of SARS-CoV-2 Spike protein from vaccine obtained
Posted: 11 August 2020 | Victoria Rees (Drug Target Review) | 1 comment
The SARS-CoV-2 Spike protein from a COVID-19 vaccine candidate has been characterised by researchers, supporting the neutralising antibody response it elicits.
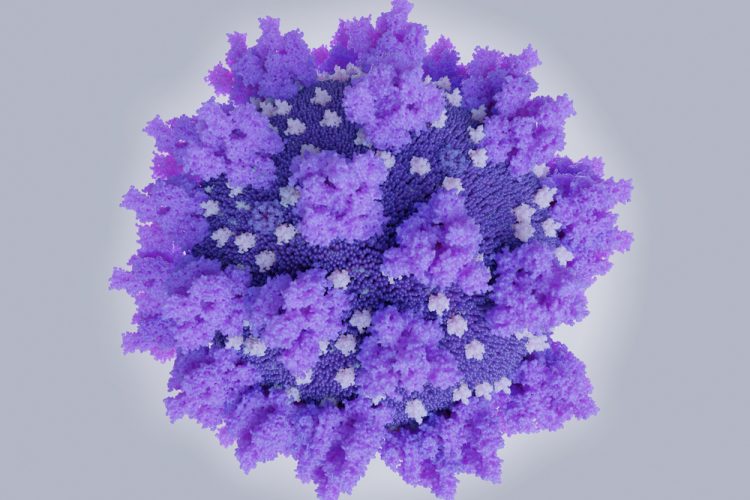

Scientists have obtained high-resolution, atomic-scale details of the structure of a SARS-CoV-2 Spike (S) protein from an experimental COVID-19 vaccine named NVX-CoV2373. According to the researchers, these details are consistent with the robust neutralising antibody responses the vaccine elicited in pre-clinical and Phase I clinical studies.
From Scripps Research, US, the researchers investigated NVX-CoV2373, which is being developed and tested in clinical trials by Novavax Inc. It uses lab-grown copies of the SARS-CoV-2 S protein to elicit neutralising antibody responses to the viral protein, with the goal of preventing new COVID-19 infections.
The structural details were acquired using low-temperature electron microscope techniques. The researchers say the molecular map of the Novavax S protein could be helpful in evaluating the vaccine’s ongoing human clinical trial results.
Biomarkers aren’t just supporting drug discovery – they’re driving it
FREE market report
From smarter trials to faster insights, this report unpacks the science, strategy and real-world impact behind the next generation of precision therapies.
What you’ll unlock:
- How biomarkers are guiding dose selection and early efficacy decisions in complex trials
- Why multi-omics, liquid biopsy and digital tools are redefining the discovery process
- What makes lab data regulatory-ready and why alignment matters from day one
Explore how biomarkers are shaping early drug development
Access the full report – it’s free!
“Our findings suggest that the speedy design and manufacture of this vaccine did not compromise the quality of the S protein structure,” said the study’s senior author Dr Andrew Ward, a professor in the Department of Integrative Structural and Computational Biology at Scripps Research.
The team say that the S protein has a floppy, ever-changing shape, which may be a strategy for evading immune recognition. The immunogen S protein uses mutations to stabilise the S protein in its functional shape so that the immune system can learn to recognise and attack it. The study demonstrated that the mutations stabilise the S protein in the desired conformation.
Ward said that the structural details also indicate that the S protein has previously undiscovered “pockets” where certain compounds could bind. “These are places where, in principle, you could target the S protein with small-molecule drugs,” he noted.
The researchers also say the coronavirus S proteins naturally form three-copy structures called trimers on virus particles and function in that trimer shape. Ward and his team, in the new study, found that the S proteins not only form trimers successfully, but can also form clusters of two or three trimers.
According to the team the specific molecular nature of interactions points to the possibility – which they can now follow up in further studies – that clustering of S trimers occurs naturally on SARS-CoV-2 to enhance its infectivity.
“It’s possible that the virus needs more than one S trimer to pierce a host cell, so this tendency to form higher-order clusters may underlie its infectiousness,” Ward said.
The pre-print results of the study can be found in bioRxiv.
Related topics
Antibodies, Drug Development, Microscopy, Protein, Proteomics, Research & Development, Target Validation, Vaccine
Related conditions
Coronavirus, Covid-19
Related organisations
Scripps Research
Related people
Dr Andrew Ward








Nice answer back in return of this question with solid arguments
and describing the whole thing about that.