RNA structures of SARS-CoV-2 reveal potential drug targets
Posted: 11 November 2020 | Victoria Rees (Drug Target Review) | No comments yet
The SARS-CoV-2 RNA genome structure has been studied by researchers who identified several potential drug targets.

The SARS-CoV-2 RNA genome structure has been studied in detail by researchers, revealing potential targets for the development of drugs against the virus. The scientists say that this is the first time that the structure of the entire novel coronavirus RNA (one of the longest viral RNAs with approximately 30,000 nucleotides) was determined.
The study was conducted at the University of Groningen and Leiden University, both Netherlands and the International Institute of Molecular and Cell Biology, Poland.
According to the researchers, SARS-CoV-2 is a beta coronavirus with a linear single-stranded, positive-sense RNA genome. Similar to those in other RNA viruses, the SARS-CoV-2 RNA structures are expected to play a crucial role in how the coronavirus replicates in human cells. Despite this importance, only a handful of functionally relevant coronavirus structural RNA elements have been studied to date. This prompted the researchers to perform an extensive characterisation of the SARS-CoV-2 RNA genome structure.
The team probed the RNA structure to obtain single-base resolution secondary structure maps of the full SARS-CoV-2 coronavirus genome both in vitro and in living infected cells. Subsequently, they identified at least 87 regions in the SARS-CoV-2 RNA sequence that appear to form well-defined compact structures. Of these, at least 10 percent are under strong evolutionary selection pressure among coronaviruses, suggesting functional relevance.
“We first identified the structures in vitro and subsequently confirmed their presence in the RNA of viruses inside cells,” explained study co-ordinator Dr Danny Incarnato. “This means that our results are very robust… Furthermore, a number of the structures are conserved between different coronaviruses, meaning that a successful drug targeting SARS-CoV-2 could also be effective against future new virus strains.”
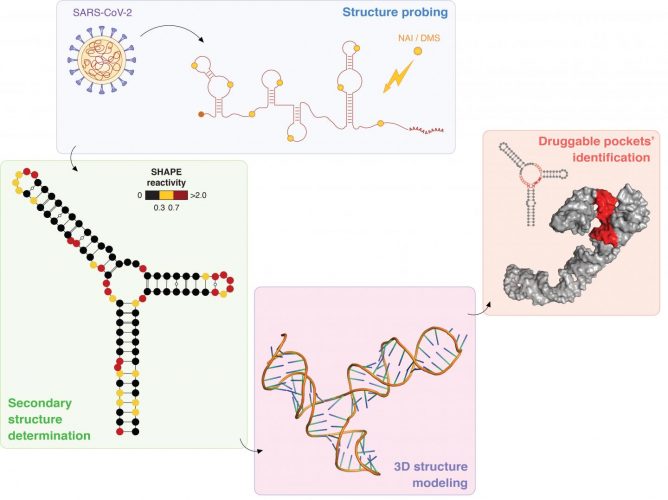

Schematic representation of the experimental approach adopted by the researchers. After ‘probing’ the structure of the SARS-CoV-2 RNA genome using specific chemical compounds, the secondary structure of stable RNA elements throughout the viral genome has been determined. Using this information, the three-dimensional (3D) model of these RNA structure elements has been computationally determined, allowing the identification of potential pockets that might provide targets for the development of antiviral therapeutic strategies [credit: Danny Incarnato, University of Groningen].
Also, pockets were identified in some RNA structures that could be targeted by small molecules to hamper the function of the viral RNA. The scientists also identified parts of the SARS-CoV-2 RNA that are intrinsically unstructured.
“These could be targeted by antisense oligonucleotide therapeutics,” commented Incarnato. Adding short nucleic acid strands that can bind to these viral RNA sections would create double-stranded regions, which are naturally targeted by enzymes inside human cells.
The researchers say that this collaborative study establishes a firm foundation for future work aimed at developing potential small-molecule drugs for the treatment of SARS-CoV-2 infections and possibly also infections by other coronaviruses.
The results were published in Nucleic Acid Research.
Related topics
Disease Research, Drug Targets, Genetic Analysis, Genomics, Research & Development, RNAs
Related conditions
Covid-19
Related organisations
International Institute of Molecular and Cell Biology, Leiden University, University of Groningen
Related people
Dr Danny Incarnato