Versatile applications of next-generation sequencing in pharmaceutics
Posted: 4 December 2016 | Pushpanathan Muthuirulan (National Institutes of Health) | No comments yet
Next-generation sequencing (NGS) technologies hold great promise for unravelling the mysteries of biological systems, with the advent of DNA sequencing methods having greatly accelerated biomedical research, gaining broad applicability in disease diagnosis and therapeutics in recent years…


NGS has demonstrated the capability of producing massive amounts of sequencing data at unprecedented speed and a more affordable price for biomedical applications, which has opened up new frontiers of genomics research with exciting possibilities for disease diagnosis and therapeutic intervention. Furthermore, increased availability of innovative NGS platforms along with improved high-throughput NGS data analysis tools and computing strategies have leveraged the power of exploring NGS data information for research and clinical applications.
This article outlines how NGS technologies have been widely implemented to discern actionable genomic information for the following areas: drug screening; novel drug target identification; profiling adverse drug effects; biomarker discovery; biopharmaceutics development; vaccine production; pharmacogenomics; personalised medicine; polypharmacology and pharmacoepidemiology (see Figure 1).
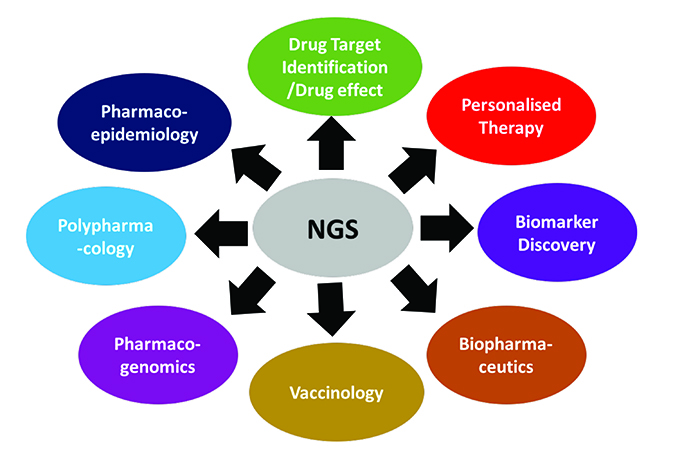

Figure 1: Versatile application of NGS technology in pharmaceutics
Introduction
DNA sequencing technologies allow us to determine the precise position of nucleotides in DNA molecules, a technique that has become instrumental in basic biological research. Rapid sequencing processes attained using technology such as NGS have become instrumental in revealing the complexity of genome architecture in several species including human, animals, plants and microbes1. The diversity and rapid evolution of innovative NGS platforms and high throughput data analysis tools have together enabled genomes, transcriptomes and interactomes to be comprehensively analysed, providing biologically meaningful and clinically relevant information for drug discovery and development2. NGS technology not only provides insight into basic research, but also offers immediate application benefits.
Scientists across the world in many different fields are utilising these technologies for improved diagnostics, prognostics and targeted therapies for cancer and other infectious diseases. Large-scale studies involving genomics, exomics, metagenomics, epigenomics and transcriptomics have now become a reality3.
Drug discovery and development
Target identification is one of the most important steps in the drug discovery process. Several methods have been used to identify and validate potential drug targets, with the ultimate aim being to assess potential therapeutic benefits associated with the modification of proposed drug targets in patients participating in clinical trials. In this context NGS can be used to drive and direct the development of novel therapeutic agents that affect the biological targets associated with disease progression4.
NGS also provides a powerful platform for identifying disease-associated mutations, enabling patients’ diseases to be more accurately diagnosed by providing comprehensive genome sequence information. In the case of antibiotics and chemotherapeutics, this enables drug response and the mechanism of drug resistance to be determined, so that the correct therapeutic intervention can be determined5. An individual’s response to therapeutic agents can be extensively analysed by large-scale screening of single nucleotide polymorphisms (SNPs) using NGS technology. Exome sequencing followed by interpretation based on bioinformatics analysis of sequencing data can effectively identify specific genes associated with the patient’s clinical conditions6. This clinically relevant information allows specific drugs for rare diseases that have no existing treatments to be designed.
During recent years, DNA-encoded chemical libraries have been used in drug discovery research. These libraries consist of DNA barcodes with unique sequences that, when sequenced, facilitate the identification of the small molecules bound to immobilised specific target molecules7.
Biomarker discovery
Over recent decades biomarker discovery has become instrumental in many aspects of biomedical research and today has become the foundation for personalised medicine, greatly improving medical diagnostics procedures for detecting and predicting diseases. Large-scale data generation using NGS and unbiased bioinformatics data analysis has identified novel biomarkers that have made the drug discovery process much quicker and more efficient. Cancer is the most common human genetic disease that arises from gene mutations as well as changes in epigenetic and transcriptional profiles. These genetic changes may serve as valuable and potential biomarkers for early diagnosis, staging, and in-depth molecular characterisation of cancer for personalised therapy8.
Quantification of microRNAs, particularly small ncRNAs (sncRNAs), offers attractive hope as biomarkers of diseases such as cancer, liver disease, cardiovascular disease and central nervous system disorders. Thus, high-throughput sequencing quantification using NGS generates valuable information that can contribute to our knowledge of sncRNAs as potential biomarkers and mediators of biological functions and diseases9. Biomarker identification using NGS also offers genetic diversity clarification for pathogen characterisation and surveillance10.
Biopharmaceutics development
Early applications of NGS focused on the therapeutic areas of oncology and infectious diseases. However, recent applications have broadened the knowledge of natural antibody repertoires (antibodyomes) and their evolution in response to microbial infections. For example, NGS data can provide a clear understanding of antibody diversity, which provides effective pathways to engineer antibodies that specifically target desired antigens for widespread therapeutic application11. Deep sequencing combined with proteomic techniques might provide a better picture of the mechanism of the humoral response, antibody clonal selection and the structure and chemical nature of the binding landscape of antibodies. These would, in turn, enable researchers to generate designer libraries that would specifically address certain classes of antigens and thus improve biopharmaceutics development12.
During recent years, deep DNA sequencing strategies have also been used to characterise cDNA phage display libraries and can aid in screening the protein-protein interactions associated with various disease pathologies13. Thus, incorporation of NGS technology into the area of pharmaceutics will aid the identification of new targets that warrant large-scale prospective investigation as well as improve the understanding of cross-talk between activated signalling pathways associated with disease conditions. This, in turn, may open up the door to more successful drug development.
Vaccine development
Research into vaccines increasingly involves NGS methods. Combinations of studies involving both pathogen and host are the mainstay of vaccine development and NGS has great transformative potential in the field of vaccinology by virtue of its versatile applications. These technologies are helping unravel the complex dynamics of pathogen evolution and host responses to the pathogen, thus substantially improving the likelihood that successful vaccines are developed14.
Transcriptome sequencing offers comprehensive gene expression profiling of target populations and has led to the identification and characterisation of differences in transcript abundance at genetic level, essentially providing an overview of the transcribed product in response to vaccination. Being able to compare transcriptome profiles between ‘good’ and ‘poor’ responders to vaccines might aid the generation of signature immune-response profiles that could enable prediction adverse responses to the vaccine as well as help in the understanding of genotype-phenotype associations15. Thus, NGS technology offers a way to develop safer vaccines and in the future is likely to take vaccinology into the era of pre-vaccination genome-wide screening to generate a ‘personalised vaccination regimen’ for reducing the rates of failures and adverse events.

Research into vaccines increasingly involves NGS methods
Pharmacogenomics and personalised medicine
Pharmacogenomics, which involves the development of strategies for personalised therapy to optimise drug effectiveness and minimise toxicity, has shed light on the role of genes and genomic variants in humans. The advent of NGS has provided unprecedented opportunities for analysing whole genome or exome (coding regions of DNA) sequencing data which, in turn, helps in detecting individuals’ variomes that influence drug response16. In clinical settings, pharmacogenomics tests using NGS have enabled clinicians to predict a patient’s response to drug therapy based on their individual genetic makeup. NGS can comprehensively screen well-established pharmacogenomics genes in a single, cost effective test that provides medically relevant data that aids clinical decision-making17.
Overall, NGS assists the development of drugs by maximising their therapeutic effects, offering new ways for designing therapeutic strategies based on individual genetic makeup and enabling the risk factors associated with drugs to be determined18. There are now opportunities to develop personalised therapies using NGS tools, since we are able to understand genetic variations between people and thus provide more optimal targeted disease therapy.
Polypharmacology
Polypharmacology is the use of pharmaceutical agents that act on multiple targets pertaining to unique or multiple disease pathways. During recent years, polypharmacology has become recognised as a valuable tool for drug discovery and development. NGS enables multiple targets pertaining to disease pathways to be discovered, thereby providing an effective path for developing drug molecules that can typically bind multiple targets18.
The combination of computational design and chemical synthesis of libraries of ligands identified through NGS analysis could provide an alternative way to more effectively obtain bioactive agents with precise on- and off-target binding19. NGS technology enables predictions to be made based on ligand/targets, genotype and phenotype. Furthermore, NGS can be utilised in studying the drug domain network, which allow us to explore the role of protein domains as drug targets and accelerates polyphamacological research by characterising privileged sets of druggable domains20.
Pharmacoepidemiology
Continuously monitoring undesirable effects and other safety concerns arising from drugs in well-defined populations is known as pharmacoepidemiology. In such studies, NGS has enabled researchers to understand the determinants of beneficial and adverse drug effects; the impact of genetic variation on drug effect; dose-response relationships of drugs; and clinical effects of drug-drug interactions21. Implementating NGS technology in pharmacoepidemiology might provide a valuable source of information for monitoring adverse drug reactions (ADRs) and assessing the benefit-risk profile of drugs. It could also increase awareness among healthcare professionals on the importance of ADRs. Thus, NGS provides a more informative profile of drug products that could benefit clinicians in promoting human healthcare.
Conclusion
NGS is an extremely powerful tool for conducting basic biological research and it offers unmatched opportunities for improved diagnostics, prognostics and targeted therapies. The application of NGS in the translation of research findings from bedside to clinic is ever increasing. This is necessitating the development of computational tools to manage the growing wealth of sequencing data that it produces. In turn, the efficient analysis and interpretation of enormous databases of information drives and directs the development of safer drugs with greater efficacy and less toxicity, so improving patients’ health and preventing diseases.
Biography


References
- Levy SE, Myers RM. Advancements in Next-Generation Sequencing. Annu. Rev. Genomics Hum. Genet. 2016; 17:95–115
- Shendure J, Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008; 26:1135–1145
- Chee‐Seng K, En Yun L, Yudi P, Kee‐Seng C. Next generation sequencing technologies and their applications. eLS. 2010
- Beltran H, Yelensky R, Frampton GM, Park K, Downing SR, MacDonald TY, et al. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur. Urol. 2013; 63:920–926
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008; 321:956–960
- Voisey J, Morris CP. SNP technologies for drug discovery: a current review. Drug Discov. Technol. 2008; 5:230–235
- Ding Y, O’Keefe H, DeLorey JL, Israel DI, Messer JA, Chiu CH, et al. Discovery of potent and selective inhibitors for ADAMTS-4 through DNA-encoded library technology (ELT). ACS Med. Chem. Lett. 2015; 6:888–893
- Desai AN, Jere A. Next-Generation Sequencing for Cancer Biomarker Discovery. In Next Generation Sequencing in Cancer Research, Springer International Publishing. 2015; 2:103–125
- Lopez JP, Cruceanu C, Fiori LM, Laboissiere S, Guillet I, Fontaine J, et al. Biomarker discovery: quantification of microRNAs and other small non-coding RNAs using next generation sequencing. BMC Med. Genomics, 2015 8:1
- Zhao W, Chen JJ, Foley S, Wang Y, Zhao S, Basinger J, Zou W. Biomarker identification from next-generation sequencing data for pathogen bacteria characterization and surveillance. Med. 2015; 9:1253–1264
- Woollard PM, Mehta NA, Vamathevan, JJ, Van Horn S, Bonde BK, Dow DJ. The application of next-generation sequencing technologies to drug discovery and development. Drug discov. Today, 2011; 16:512–519
- Mathonet P, Ullman C. The application of next generation sequencing to the understanding of antibody repertoires. Immunol. 2013; 4: 265
- Di Niro R, Sulic AM, Mignone F, D’Angelo S, Bordoni R, Iacono M. Rapid interactome profiling by massive sequencing. Nucleic Acids Res.2010; 38: e110-e110
- Luciani F, Bull RA, Lloyd AR. Next generation deep sequencing and vaccine design: today and tomorrow. Trends Biotechnol. 2012; 30: 443–452
- Dhiman N, Smith DI, Poland, GA. Next-generation sequencing: a transformative tool for vaccinology. Expert Rev. Vaccines, 2009; 8:963–967
- Katsila T, Patrinos GP. Whole genome sequencing in pharmacogenomics. Pharmacol, 2015; 6: 61
- Rabbani B, Nakaoka H, Akhondzadeh S, Tekin M, Mahdieh N. Next generation sequencing: implications in personalized medicine and pharmacogenomics. BioSyst.2016; 12:1818–1830
- Nawab DH. The Pharmaceutical Applications of Next Generation Sequencing in Oncology Drug Designing and Development. Next Generat. Sequenc & Applic, 2015. 2:116. doi:10.4172/jngsa.1000116
- Rastelli G, Pinzi L. Computational polypharmacology comes of age. Pharmacol. 2015: 6
- Moya-García AA, Ranea JA. Insights into polypharmacology from drug-domain associations. Bioinformatics, 2013; 29:1934–1937
- Freedman AN, Sansbury LB, Figg WD, Potosky AL, Smith SRW, Khoury MJ, et al. Cancer pharmacogenomics and pharmacoepidemiology: setting a research agenda to accelerate translation. Natl Cancer Inst. 2010; 102:1698–705