Development of a physiologically relevant lung model for understanding SARS-CoV-2 infection
Posted: 27 November 2020 | Professor Lyle Armstrong (Newcastle University and Newcells Biotech) | No comments yet
COVID-19 is known to infect the lungs; however, the dynamics of viral infection and replication are poorly understood. Alongside the Liverpool School of Tropical Medicine, Professor Lyle Armstrong and colleagues have been working to develop a human lung epithelium model of SARS-CoV-2 infection and replication. In this article, he describes data that show the model can be successfully infected with SARS‑CoV-2 via angiotensin converting enzyme 2 (ACE2) proteins expressed on the cell surface and allows replication of the virus once inside its cells. The model provides a valuable platform to study SARS‑CoV-2 pathogenesis and evaluate candidate vaccines and therapeutics.

Characteristics of SARS-CoV-2
In December 2019, new unexplained cases of pneumonia and respiratory distress were reported by officials in Wuhan, China.1 In January 2020, the pathogen was identified as a coronavirus similar to the pathogen that caused previous pandemics such as Middle East Respiratory Syndrome coronavirus (MERS-CoV) and Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) and was therefore named SARS-CoV-2.2 Coronaviruses are a large family of viruses, several of which are known to infect humans, causing primarily respiratory tract infections. The first four (229E, NL63, O
C43 and HKU1) generally produce milder symptoms, whereas MERS-CoV, SARS-CoV and SARS-CoV-2 are more life-threatening. This has proven to be particularly true of the latter, since it spread so quickly that the World Health Organization (WHO) declared coronavirus disease 2019 (COVID-19) a public health emergency on 30 January 2020 and subsequently a pandemic on 11 March 2020. At the time of writing, more than 34 million cases have been reported across 188 countries and territories, with the number of deaths exceeding one million.3

Coronaviruses (CoVs) belong to the Coronaviridae family of order Nidovirales. They are classified into four gen
era; Alpha-, Beta-, Gamma- or Deltacoronavirus. The CoVs genome is 26‑32 kilobases long, which is the largest RNA genome reported to date.4 The CoV particles are spherical in shape, with diameters ranging between 50 and 200nm. Like all viruses, these pathogens can only complete their replication cycle by parasitising other cells. This is accomplished by viral attachment to specific proteins on the cell surface, followed by transit of the viral genome across the cell membrane and the subsequent synthesis of all the components needed to build multiple new viral particles. The surface antigens targeted by SARS-CoV-2 include the receptor for angiotensin converting enzyme-2 (ACE2R) and the matrix metalloproteinase inducer
protein, CD147.5 For the CoV genome to enter the host cell, the S1 unit of the Spike (S) protein needs to bind the receptor, followed by S protein priming by host proteases, which enables fusion of the viral and cellular membranes. A recent publication has shown that S priming is carried out by transmembrane protease serine type 2 (TMPRSS2).6
Since ACE2R and TMPRSS2 are expressed by a wide range of cells, the virus has many potential targets in the human body. SARS-CoV-2 has been shown to attack the respiratory system, gastrointestinal system, heart, kidney, liver and central nervous system leading to multiple organ failure.7 Previous studies have shown that rapid viral replication in epithelial cells, vascular endothelial cells and macrophages can lead to massive epithelial and endothelial cell apoptosis causing the release of pro-inflammatory cytokines,8-10 which can potentially cause lung damage and diminished patient survival. This is exemplified by the observation that in SARS-CoV-2 infected individuals, interleukin (IL)-6, IL-10 and TNFα surge during illness and decline during recovery. Severely affected patients who require intensive care treatment can be distinguished by significantly higher levels of IL-6, IL-10 and TNFα and fewer CD4+ and CD8+ T cells.11 Although it is likely that ingress of large numbers of cytokine secreting inflammatory macrophages into the lung tissue accounts for a significant proportion of the cytokines detected in such cases, the initial damage to the lung epithelial and endothelial cells probably contributes not only to the overall concentration of cytokines, but may also be responsible for recruitment of inflammatory macrophages. Other mechanisms besides apoptosis can lead to activation of the inflammasome. The binding of SARS-CoV-2 to the Toll-like receptor causes release of pro-IL-1β which is cleaved by caspase-1, followed by inflammasome activation and production of active mature IL-1β which is a mediator of lung inflammation, fever and fibrosis.
Development of a structurally representative, virally infected model for pre-clinical testing
Despite this understanding of the mechanisms by which SARS-CoV type viruses damage the lung epithelia and vascular endothelia, effective treatments for COVID-19 are lacking. Therefore, there is an urgent need for pre-clinical tools that may contribute to the development of COVID-19 therapies.
We have used our proprietary method to generate a purified induced pluripotent stem cell (iPSC)-derived basal cell population that can be expanded over multiple passages, while retaining their differentiation potential. The purified population can be differentiated into a stratified epithelium with a striking resemblance to in vivo human airway. The cells can differentiate into beating cilia and functional secretory cells (goblet and club cells), which form a protective mucus layer.
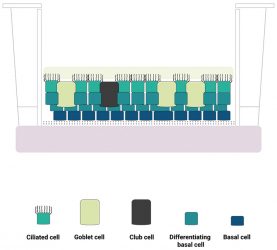

Figure 1: A schematic diagram of the human lung epithelium model developed by Newcells Biotech and the Liverpool School of Tropical Medicine. The purified induced pluripotent stem cell (iPSC)‑derived basal cell population is differentiated into a stratified epithelium representative of the in vivo human airway. We have confirmed the presence of functional basal cells, goblet cells, club cells, ciliated cells and an apical mucus layer with beating cilia.
Such “airway equivalents” contain ciliated epithelial cells, capable of forming tight junctions between themselves, club cells, which are required for maintenance of epithelial integrity and repair,12 and ΔNp63 positive basal cells, located on the bottom layer of the construct. Mucin1 expression indicates the presence of mucous producing goblet cells. Importantly, expression of the angiotensin converting enzyme receptor is detected at a higher level than observed in airway equivalents constructed from primary basal cells isolated from ex vivo airway tissue samples. Generation of lung organoids from pluripotent stem cells has been reported by other groups; however, the format in which individual cells are presented in many of these structures differs greatly from the architecture of the airway epithelia in vivo. Many organoid systems adopt a spheroidal architecture13-15 and may not present a mucous layer barrier, which reduces their utility as viral infection models. Three-dimensional (3D) airway equivalents can be generated from basal cells isolated ex vivo, but these are subject to batch and inter-individual variability, which complicates interpretation of data. Basal cells and airway equivalents can be manufactured economically in large numbers from iPSCs with controllable batch variability and a common genetic background. Thus, we believe that this model is advantageous as a pre-clinical tool for developing therapies to treat COVID-19.
The fact that the lung constructs will undergo infection by and support replication of SARS-CoV-2 underlines this utility. Working with the Liverpool School of Tropical Medicine, we have shown the release of live virions for up to 96 hours after infection, with the virus at a multiplicity of infection value of 0.01 (eg, one virus for every 100 cells in the lung construct). Moreover, the constructs secrete cytokines in a manner similar to the lungs of COVID-19 patients.
Using the model to investigate replication, dose response and viral replication

The iPSC-derived lung constructs are available on transwell inserts (ThinCerts™, Greiner bio-one) with PneumaCult culture medium supplemented with heparin and hydrocortisone added up to the basolateral face of the membrane. This ensures that the airway epithelium is kept at an air-liquid interface, which is essential for the function of the product. When the plates arrive in the laboratory, a medium change is advised followed by a 24‑hour recovery before any experimental studies can begin.
Applications of the model will of course be dependent on the scientific questions which investigators seek to answer, but the search for inhibitors of SARS-CoV-2 replication is a relevant and timely use. Constructs can be infected at various multiplicities of infection and the release of virions into the air-liquid interface can be quantified by adding samples of media collected from the apical surface into analyses such as the plaque assay (in which cells capable of infection are exposed to the viral samples and areas of dead cells are measured) or quantitative polymerase chain reaction (qPCR) amplification of the viral RNA genome. These assays can be performed over a range of concentrations of prospective antiviral agents, so that the level of replication inhibition can be measured as a dose‑response curve.
About the author
Professor Lyle Armstrong is Chief Scientific Officer at Newcells Biotech and Professor of Stem Cell Sciences at Newcastle University. His focus is on new methods to reprogramme the cells found in the human body so that they can be converted into medically useful cells. He works with experts in production of induced pluripotent stem cells (iPSCs) and their differentiation into a wide range of reliable and reproducible cell and tissue types.
References
- Zhu, N. Zhang, D. Wang, W. Li, X. Yang, B. Song, J. Zhao, X. Huang, B. Shi, W. Lu, R. Niu, P. Zhan, F. Ma, X. Wang, D. Xu, W. Wu, G. Gao, G.F. Tan, W. China Novel Coronavirus Investigating and Research Team. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733;
- World Health Organisation (WHO) Naming the coronavirus disease (COVID-19) and the virus that causes it. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it
- Woldometer. COVID-19 Coronavirus Pandemic. Available from: https://www.worldometers.info/coronavirus/ [Accessed 30th September 2020]
- Lee, H.J. Shieh, C.K. Gorbalenya, A.E. Koonin, E.V. La Monica, N. Tuler, J. Bagdzhadzhyan, A. Lai, M.M. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180(2):567-82
- Ou, X. Liu, Y. Lei, X. Li, P. Mi, D. Ren, L. Guo, L. Guo, R. Chen, T. Hu, J. Xiang, Z. Mu, Z. Chen, X. Chen, J. Hu, K. Jin, Q. Wang, J. Qian, Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11(1):1620
- Hoffmann, M. Kleine-Weber, H. Schroeder, S. Krüger, N. Herrler, T. Erichsen, S. Schiergens, T.S. Herrler, G. Wu, N-H. Nitsche, A. Müller, M.A. Drosten, C. Pöhlmann, S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271-280
- Su, S. Wong, G. Shi, W. Liu, J. Lai, A.C.K. Zhou, J. Liu, W. Bi, Y. Gao, G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016;24(6):490− 502
- Fu, Y. Cheng, Y. Wu, Y. Understanding SARS-CoV-2-Mediated Inflammatory Responses: From Mechanisms to Potential Therapeutic Tools. Virol. Sin. 2020; 35(3):266-271
- Sarzi-Puttini, P. Giorgi, V. Sirotti, S. Marotto, D. Ardizzone, S. Rizzardini, G. Antinori, S. Galli, M. COVID-19, cytokines and immunosuppression: what can we learn from severe acute respiratory syndrome? Clin. Exp. Rheumatol. 2020;38:337-342.
- Conti, P. Ronconi, G. Caraffa, A. Gallenga, C.E. Ross, R. Frydas, I. Kritas, S.K. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents. 2020;34(2):327-331
- Diao, B. Wang, C. Tan, Y. Chen, X. Liu, Y. Ning, L. Chen, L. Li, M. Liu, Y. Wang, G. Yuan, Z. Feng, Z. Wu, Y. Chen, Y. Reduction and Functional Exhaustion of T Cells in Patients with Coronavirus Disease 2019 (COVID-19). medRxiv. 2020;https://doi.org/10.1101/2020.02.18.20024364
- Wong, A.P. Keating, A. Waddell, T.K. Airway regeneration: the role of the Clara cell secretory protein and the cells that express it. Cytotherapy. 2009;11(6):676–87
- Shi, R. Radulovich, N. Ng, C. Liu, N. Notsuda, H. Cabanero, M. Martins-Filho, S.N. Raghavan, V. Li, Q. Mer, A.S. Rosen, J.C. Li, M. Wang, Y-H. Tamblyn, L. Pham, N-A. Haibe-Kains, B. Liu, G. Moghal, N. Tsao, M-S. Organoid Cultures as Preclinical Models of Non–Small Cell Lung Cancer. Clin. Cancer Res. 2020;26(5):1162-1174
- Leibel, S.L. Winquist, A. Tseu, I. Wang, J. Luo, D. Shojaie, S. Nathan, N. Snyder, E. Post, M. Reversal of Surfactant Protein B Deficiency in Patient Specific Human Induced Pluripotent Stem Cell Derived Lung Organoids by Gene Therapy. Sci. Rep. 2019;9(1):13450
- Strikoudis, A. Cieślak, A. Loffredo, L. Chen, Y-W. Patel, N. Saqi, A. Lederer, D. J. Snoeck, H-W. Modeling of Fibrotic Lung Disease Using 3D Organoids Derived from Human Pluripotent Stem Cells. Cell Rep. 2019;27(12):3709-3723
Related topics
Cell Cultures, Cytokines, Disease Research, Drug Development, Drug Targets, Immunology, In Vitro, Organoids, Screening
Related conditions
Coronavirus, Covid-19, Middle East respiratory syndrome coronavirus (MERS-CoV), Severe Acute Respiratory Syndrome (SARS)
Related organisations
Liverpool School of Tropical Medicine (LSTM), World Health Organization (WHO)