Five SARS-CoV-2 antiviral candidates revealed in drug screening study
Posted: 9 February 2022 | Ria Kakkad (Drug Target Review) | No comments yet
Using virtual screening, researchers have discovered several natural compounds that could inhibit the SARS-CoV-2 main protease.
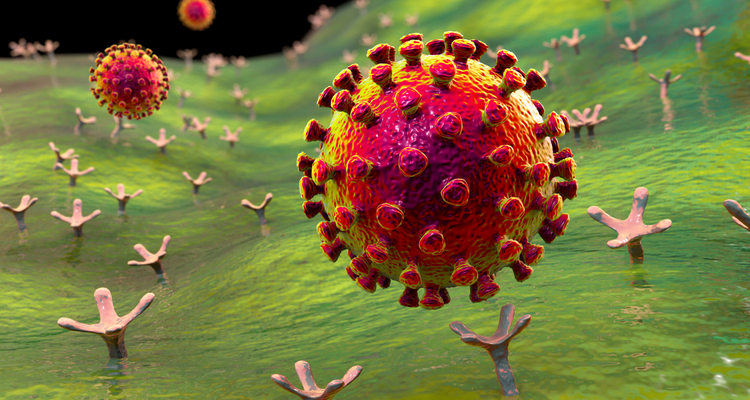
A study conducted at the University of Barcelona, Spain has revealed natural products that can inhibit Mpro, a non-structural protein and main protease of SARS-CoV-2, which is essential in the replication and transcription of the virus. It is therefore regarded as a potential therapeutic target, as it could prevent the virus from progressing.
This new scientific advance is based on the use of in silico virtual screening, a computational analytical technique for selecting molecules that are candidates to future drugs in a chemical library [credit: University of Barcelona].
Published in the Journal of Chemical Information and Modelling, the new research establishes potential novel therapeutic strategies to fight COVID-19. Based on in silico virtual screening, the new scientific advance enabled a computational analysis of molecules placed in databases to identify and select compounds that are of potential pharmacological interest against SARS-CoV-2. The researchers say that this is a reliable, fast and efficient way of finding bioactive compounds of large chemical collections against a specific molecular target.
The study consisted of a virtual screening of a natural product database, a chemical library containing nearly 2,000 compounds. Using this, the research team built series of conformations of the SARS-CoV-2 Mpro characterised with a molecular dynamics study. This showed the Mpro protease in its apo shape through conventional simulations (cMD) and Gaussian accelerated molecular dynamics (GaMD) simulations and it defined a series of representative structures.
The research team were then able to use these structures to carry out ensemble docking, a bioinformatic method that allows the prediction and calculation using computational techniques of the most favourable position of interaction between a ligand and its target protein. Then, they carried out an iterative procedure which increased the length of cMD of protein-ligand complexes and calculated the free binding energy to select the most promising candidates. Eleven compounds were selected from these results and were tested in vitro for their ability to contain Mpro protease. Five antiviral candidates were subsequently identified as promising inhibitors of Mpro protease.
Related topics
Drug Development, Drug Discovery, Drug Discovery Processes, Drug Leads, Pharmacology, Screening
Related conditions
Coronavirus, Covid-19
Related organisations
University of Barcelona