A new technique to unravel gene regulation
Posted: 4 April 2022 | Ria Kakkad (Drug Target Review) | No comments yet
Researchers at Hubrecht Institute, Netherlands have developed a technique to measure both gene expression and DNA packaging at the same time.

Researchers at Hubrecht Institute, Netherlands have developed a technique to measure both gene expression and DNA packaging at the same time. The method, called EpiDamID, determines the location of modified proteins around which the DNA is wrapped. It is important to gather information about these modifications as they influence the accessibility of DNA, therefore affecting the gene activity. EpiDamID is therefore valuable for research into the early development of organisms. The study was recently published in Molecular Cell.
To fit DNA into the nucleus of a cell, it is tightly packed around nuclear proteins: histones. Depending on the tightness of this winding, the DNA can be (in)accessible to other proteins. This therefore determines whether the process of gene expression, translation of DNA into RNA and eventually into proteins, can take place.
The tightness of DNA winding around histones is regulated by the addition of molecular groups, so-called post-translational modifications (PTMs), to the histones. For example, if certain molecules are added to the histones, the DNA winding is loosened. This makes the DNA more accessible for certain proteins and causes the genes in this part of the DNA to become active or expressed. Furthermore, proteins that are crucial for gene expression can directly recognise and bind the PTMs. This enables transcription, the process of DNA copying.
The regulation of gene expression, for instance through PTMs, is also known as epigenetic regulation. Since all cells in a body have the same DNA, regulation of gene expression is needed to (de)activate specific functions in individual cells. For instance, heart muscle cells have different functions than skin cells, thus require different genes to be expressed.
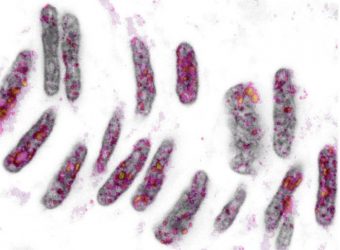

Zebrafish notochord nuclei at 15-somite stage. Grey: nuclear DNA (DAPI). Colour: histone H3K9me3
[Credit: Phong Nguyen, Franka Rang & Kim de Luca. Copyright Hubrecht Institute].
To understand how PTMs affect gene expression, the researchers designed a new method to determine the location of the modifications. With EpiDamID, researchers can analyse single cells, whereas previous methods were only able to measure a large group of cells. Analysis on such a small-scale result in knowledge on how DNA winding differs per cell, rather than information on the average DNA winding of many cells.
EpiDamID is based on DamID, a technique which is used to determine the binding location of certain DNA-binding proteins. Using EpiDamID, the binding location of specific PTMs on histone proteins can be detected in single cells. Compared to others, a great advantage of this technique is that researchers need very limited material. Furthermore, EpiDamID can be used in combination with other methods, such as microscopy, to study regulation of gene expression on different levels.
Following the development of this technique, the group will focus on the role of PTMs from the point of view of developmental biology. As single cells are analysed using EpiDamID, only a limited amount of material is needed to generate enough data. This allows researchers to study the early development of organisms from its first cell divisions, when the embryo consists of only a few cells.
Related topics
DNA, Genome Editing, Genomics, Research & Development, Technology
Related organisations
Hubrecht Institute







