A novel tool for studying autoimmune diseases related to COVID-19
Posted: 19 May 2022 | Ria Kakkad (Drug Target Review) | No comments yet
Researchers have developed a new bioinformatics pipeline that could help investigate the mechanism underlying the development of autoimmune diseases following SARS-CoV-2 infection.


Autoimmune diseases arise when the immune system confuses healthy cells with pathogens and starts attacking them. But the precise mechanism underlying this process is unknown. One of the possible mechanisms suggested to be involved is what is called “molecular mimicry,” in which an autoimmune reaction is triggered when a T-cell receptor or an antibody produced from a B-cell directed against a specific antigen bind with an autoantigen, which is an antigen produced from our own body. This occurs due to a molecular or structural resemblance between the epitopes of the antigens. However, a comprehensive investigation of the role of molecular mimicry in the development of such autoimmune diseases has not yet been performed due to the complexity of the epitope search and the lack of standardised tools.
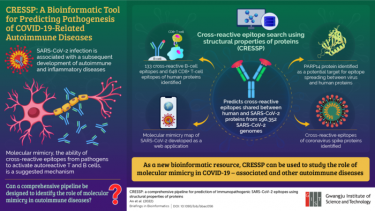
To uncover the uncertainty, a team of researchers from the Gwangju Institute of Science and Technology (GIST) have developed a new bioinformatics pipeline. Their new tool, called Cross Reactive Epitope Search Using Structural Properties of Proteins (CRESSP), was recently reported in the Briefings in Bioinformatics.
“Previous studies on molecular mimicry used bioinformatics pipelines different from one another that often involved complex algorithms and were not scalable to proteome scales. In light of this, we developed a pipeline that is easily accessible and scalable,” explained Professor Jihwan Park, research lead. “It uses the structural properties of proteins to identify epitope similarities between two proteins of interest, such as human and SARS-CoV-2 proteins.”
Automation now plays a central role in discovery. From self-driving laboratories to real-time bioprocessing
This report explores how data-driven systems improve reproducibility, speed decisions and make scale achievable across research and development.
Inside the report:
- Advance discovery through miniaturised, high-throughput and animal-free systems
- Integrate AI, robotics and analytics to speed decision-making
- Streamline cell therapy and bioprocess QC for scale and compliance
- And more!
This report unlocks perspectives that show how automation is changing the scale and quality of discovery. The result is faster insight, stronger data and better science – access your free copy today
Using CRESSP, the team screened 4,911,245 proteins from 196,352 SARS-CoV-2 genomes obtained from an open-access database. The pipeline narrowed down 133 cross-reactive B-cell and 648 CD8+ T-cell epitopes that could be responsible for COVID-related autoimmune diseases. It further identified a protein target, PARP14, to be a potential initiator of epitope spreading between COVID-19 virus and human lung proteins.

A new bioinformatics pipeline helps investigate the mechanism underlying the development of autoimmune diseases following SARS-CoV-2 infection
[Credit: Gwangju Institute of Science and Technology (GIST)]
The pipeline also predicted the cross-reactive epitopes of different coronavirus spike proteins. Moreover, the team developed an interactive web application to enable an interactive visualization of the molecular mimicry map of SARS-CoV-2. The pipeline is also available as an open-source package.
The team hopes their new tool will facilitate comparison between studies, providing a robust framework for further investigation on molecular mimicry and autoimmune diseases.
Related topics
Bioinformatics, Disease Research, Molecular Biology, Molecular Targets, Technology, Therapeutics
Related conditions
autoimmune disease
Related organisations
Gwangju Institute of Science and Technology (GIST)
Related people
Professor Jihwan Park