Mass spectrometry approach allows researchers to make precise protein measurements
Posted: 3 March 2020 | Victoria Rees (Drug Target Review) | No comments yet
A new method for weighing proteins at the atom level, called individual ion mass spectrometry, has been developed by American researchers.

A novel technology has enabled scientists to make precise measurements of proteins, down to their atoms. Developed at Northwestern University, US, the new approach is named individual ion mass spectrometry (I2MS) and can determine the exact mass of a huge range of intact proteins.
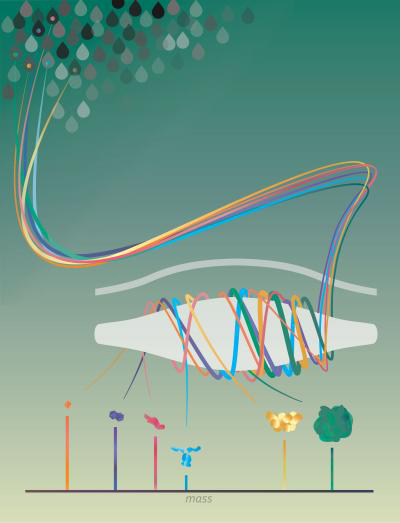

Single molecules of proteins or virus-like particles rotating around the Orbitrap analyser to determine their exact mass (credit: Northwestern University).
The researchers show their method can be used on complex mixtures of intact proteins and even whole virus particles carrying diverse cargo within them. They say this power and versatility will enable a new wave of molecular precision to be brought to diverse problems in vacinnology, virology, neurodegenerative plaques and disease biology in general. Measuring each molecule on an individual basis, the researchers say the technique could aid in the understanding of disease and infection to accelerate the design of vaccines, including the COVID-19 coronavirus.
“Quickly characterising the masses of viruses and their infectious cargo over time may help scientists understand mutations that are occurring,” said lead researcher Neil Kelleher, Walter and Mary Elizabeth Glass Chair in Life Sciences in the Weinberg College of Arts and Sciences’ departments of chemistry and molecular biosciences. “Whether directly characterising different strains of viruses or profiling different vaccine formulations, our new technology now can be deployed directly on these protein-containing samples to pursue the most urgent challenges of the day.”
The technology can help scientists to further investigate and understand the composition of the capsid (the exterior of a virus) and the infectious cargo held within the capsid, Kelleher said. As the researchers can analyse a handful of single virus particles at a time, they can extract information about precise variations in each particle.
A related study by the Kelleher group, published recently in the Journal of Proteome Research, extends analysis of I2MS to fragmentation of intact species. By fragmenting intact proteins, important information about where modifications or mutations on the protein can be identified, the researchers said. These modifications have implications for understanding how proteins change or mutate in cancer patients.
The study is published in Nature Methods.
Related topics
Analytical Techniques, Mass Spectrometry, Protein, Proteomics, Research & Development
Related organisations
Northwestern University
Related people
Neil Kelleher