Open source all-atom model of entire COVID-19 S protein released
Posted: 23 June 2020 | Victoria Rees (Drug Target Review) | No comments yet
Researchers have developed a video and model-building programme for other scientists to build full-length COVID-19 S protein models.

An international group of researchers has released the first open-source all-atom models of a full-length Spike (S) protein, which the SARS-CoV-2 virus uses to enter human cells and cause COVID-19. The researchers say this protein is of particular importance as a vaccine and antiviral drug development target.
The team was made up of researchers from Seoul National University in South Korea, University of Cambridge in the UK and Lehigh University in the US.
The scientists have created a video to illustrate how to build this membrane system from their SARS-CoV-2 S protein models. The model-building programme is open access and can be found here.
The graphical user interface the researchers used is a programme that simulates complex biomolecular systems simply, precisely and quickly. It enables scientists to understand molecular-level interactions that cannot be observed any other way.
The S protein structure was determined using cryo-EM with the receptor binding domain (RBD) up and with the RBD down. However, the researchers say this model had many missing residues. So, they first modelled the missing amino acid residues and then other missing domains. In addition, they modelled all potential glycans attached to the S protein. These glycans prevent antibody recognition, which makes it difficult to develop a vaccine. They also built a viral membrane system of an S protein for molecular dynamics simulation.
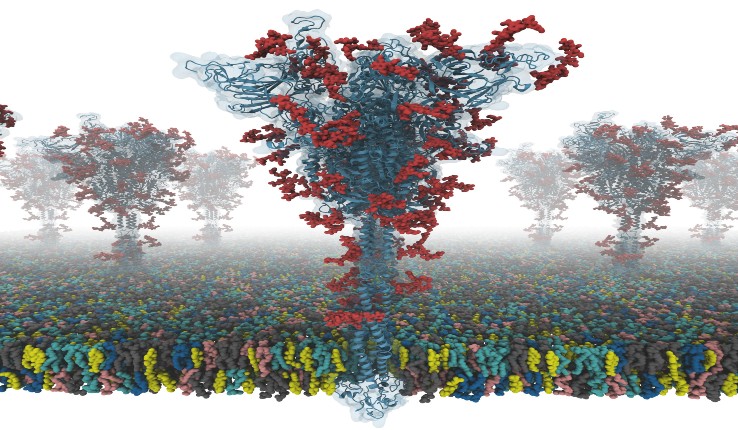

A model of an S protein [credit: Dr Yeolkyo Choi/Lehigh].
“Our models are the first fully-glycosylated full-length SARS-CoV-2 S protein models that are available to other scientists,” said Professor Wonpil Im, at Lehigh University’s Department of Biological Sciences and Bioengineering Department. “Our team spent days and nights to build these models very carefully from the known cryo-electron microscopy (cryo-EM) structure portions. Modelling was very challenging because there were many regions where simple modelling failed to provide high-quality models.”
According to the researchers, scientists can use the models to conduct innovative and novel simulation research for the prevention and treatment of COVID-19.
The team recommends reading the this pre-print paper before using any of the models. The results of the new study are published in The Journal of Physical Chemistry B.
Related topics
Disease Research, Drug Targets, Informatics, Molecular Biology, Molecular Targets, Research & Development, Structural Biology, Targets, Technology
Related conditions
Coronavirus, Covid-19
Related organisations
Lehigh University, Seoul National University, University of Cambridge
Related people
Professor Wonpil Im