Uncovering the first FDA-approved drug for Ebola virus infection
Posted: 6 February 2023 | Izzy Wood (Drug Target Review) | No comments yet
US scientists have outlined the structure and function of the first FDA approved drug for the Ebola virus.
Scientists at La Jolla Institute for Immunology (LJI), US, have uncovered the structure and function of the first Food and Drug Administration (FDA)-approved treatment for Zaire ebolavirus (Ebola virus).
Inmazeb (REGN-EB3) is a three-antibody cocktail designed to target the Ebola virus glycoprotein. The drug was first approved for clinical use in October 2020, but its exact mechanism of action has remained unclear.
In latest issue of Cell Host & Microbe, LJI researchers present a high-resolution, 3D structure of the three antibodies as they bind to the Ebola virus glycoprotein. The model reveals new information about both the drug and the virus, and how their interaction fights infection and protects against future viral mutations.
“We now know which specific amino acids the antibodies are latching onto and how their binding affects the viral glycoprotein.” said LJI President and CEO Dr Erica Ollmann Saphire.
At 3.1 angstroms, the 3D structure is the highest-resolution image of the Ebola virus surface protein ever assembled using asymmetric reconstruction. The researchers achieved this detailed view through an imaging technique called cryogenic electron microscopy (cryo-EM).
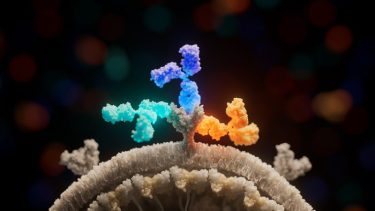
The new study shows how three antibodies (light blue, dark blue, and yellow) used in Inmazeb (REGN-EB3) bind to different regions of the Ebola virus glycoprotein (grey) to combat infection. Image credit: Ethan MacKenzie (Phospho Biomedical Animation)
“It’s like getting mugshots of a protein,” said first author Dr Vamseedhar Rayaprolu, a postdoctoral associate at LJI and now at The Pacific Northwest Centre for Cryo-EM. “We take photos of the complex that is frozen in all different angles and then stitch them together to get a 3D model.”
The imaging technique uncovered that while the overall structure of the Ebola glycoprotein has been known for some time, one region had yet to be modelled effectively—the β17-β18 loop on the protein’s glycan cap.
The team then confirmed that the drug’s three antibodies bind the glycoprotein at distinct, non-overlapping locations, maximising their effectiveness by minimising their redundancy.
Atoltivimab (REGN3470) is the specific antibody that binds the β17-β18 loop. When bound, this antibody can serve as a signal to attract the immune system, flagging infected cells to be killed via effector functions.
A second antibody, called odesivimab (REGN3471), binds to amino acids on the glycoprotein’s receptor-binding site, preventing the virus from attaching itself to human cells.
The third antibody, called maftivimab (REGN3479), binds and warps the glycoprotein’s internal fusion loop, which the virus requires to drive itself into a cell. The researchers also found evidence that maftivimab may be valuable in future therapies against other types of Ebolaviruses.
The key appears to be the maftivimab antibody. Maftivimab’s target, the viral glycoprotein’s internal fusion loop, is conserved across these Ebolaviruses. This means the loop structure has not changed significantly, even as other parts of the virus have mutated over time.
“We found that the antibodies in Inmazeb could be effective against the more closely related viruses,” added study collaborator Dr Robert Davey, Professor in the National Emerging Infectious Diseases Laboratories (NEIDL), of the Boston University Chobanian & Avedisian School of Medicine. “But for the more distantly related species, such as Marburg, more work needs to be done to devise a new antibody cocktail.”
This finding suggests that Inmazeb can provide lasting immunity against variants. The new findings may also guide the development of novel antibody drugs that target the glycoprotein more broadly or effectively.
“Knowing exactly where a drug contacts the virus helps us predict whether it is likely to still work on a new viral variant,” concluded Saphire.
“These methods and the insights from our research collaborators will be integral to the development of next-generation vaccines.”
Related topics
Antibodies, Antibody Discovery, Drug Discovery, Imaging, Targets
Related conditions
Ebola virus
Related organisations
Food and Drug Administration (FDA)
Related people
Dr Erica Ollmann Saphire, Dr Robert Davey, Dr Vamseedhar Rayaprolu