3D forces exerted by tiny cell clusters revealed through new technique
Posted: 5 March 2020 | Rachael Harper (Drug Target Review) | No comments yet
A new imaging technique, which has revealed 3D forces exerted by tiny cell clusters, could help scientists understand how tissue forms, how wounds heal or how tumours spread.
A new technique has been developed to map the three-dimensional (3D) forces that clusters of human cells exert on their surrounding environment. This could potentially help scientists better understand how tissue forms, how wounds heal or how tumours spread, the researchers said.
“We know that the way groups of cells interact with their extracellular matrix is important and we want to understand the instructions that tell these clusters to become organised into tissue-like architecture, or alternatively to become disorganised like an invasive tumour,” said Ian Y Wong, an assistant professor in Brown University’s School of Engineering, US, and corresponding author of the paper. “This technique gives us a way to profile these mechanical interactions between cells and matrix in a way that we couldn’t before.”
The new technique makes use of traction force microscopy (TFM), an imaging method that has been widely used to study the forces exerted by single cells. The goal of this new technique is to bring TFM to bear on multicellular clusters.
“We know that tumours, for example, tend to be spatially heterogeneous, with cells behaving differently throughout a tumour,” added co-first author Susan Leggett, who led this research. “So elucidating heterogenous behaviours across a multicellular cluster is something that’s important in a clinical context.”
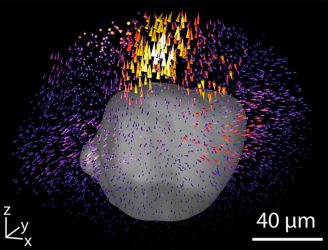
A new technique for mapping the forces that clusters of cells exert on their surroundings could be useful for studying everything from tissue development to cancer metastasis (credit: Wong Lab/Brown University).
One of the challenges when dealing with large 3D data sets is how to depict them in a convenient, quick and reader-friendly format. So Leggett and her colleagues came up with what they call Displacement Arrays of Rendered Tractions (DART), which virtually divides the volume around each cluster into 16 distinct regions. By mapping the forces that dominate each of the regions into a ‘DART-board’ display, the technique can capture the differing forces in play within a cluster in an easy-to-interpret format.
The research team streamlined the image processing required to track the fluorescent beads embedded in the biomaterial which enables the technique to image many clusters at a time, arrayed on 96-well cell culture plates. That high throughput, which had not been feasible previously, makes the technique even more powerful, the researchers say.
The team highlights that the technique could have a variety of applications, from basic biology research to clinical cancer research or precision medicine. “Basically in any setting where cells need to move in an extracellular matrix, we can use this technique to look for patterns,” said co-author Christian Franck, an associate professor of mechanical engineering at the University of Wisconsin, Madison.
This technique could also be used to study organoids, the researchers say. This approach is based on culturing primary human cells on a dish in order to screen personalised drug treatment. “You could imagine isolating patient cells from a tumour biopsy, culturing them on a 96-well plate, then treating with different drugs to see whether they affect how these cells migrate and divide,” Wong said.
The paper describing the work was published in the Proceedings of the National Academy of Sciences.
Related topics
3D printing, Cell Cultures, Imaging, Microscopy, Personalised Medicine, Screening, Targets
Related conditions
Cancer
Related organisations
Brown University
Related people
Christian Franck, Ian Y Wong, Susan Leggett