Nanobodies developed to image COVID-19 Spike protein
Posted: 20 April 2020 | Victoria Rees (Drug Target Review) | 1 comment
Researchers in the UK have selected nanobodies that bind with high affinity to the Spike protein on the COVID-19 coronavirus, enabling stabilisation for imaging.
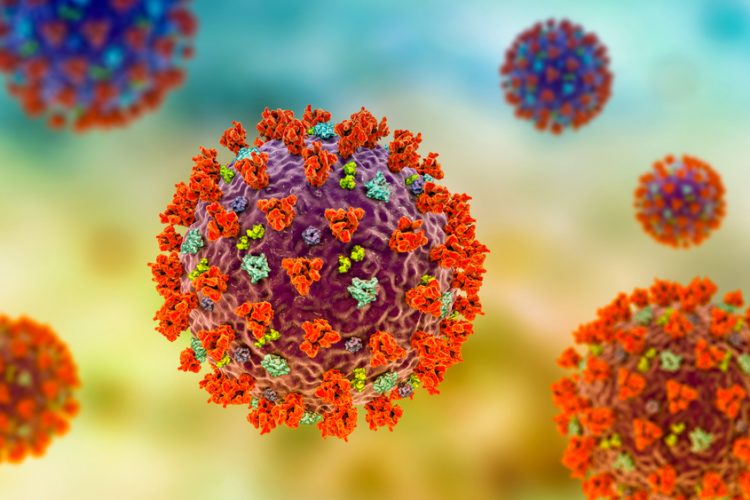

Researchers have isolated nanobodies which bind with high affinity to the Spike (S) protein of the SARS-CoV-2 virus, responsible for the COVID-19 pandemic. The scientists say their development could enable improved imaging of the coronavirus.
The team from Protein Production UK, a collaborative project led by The Rosalind Franklin Institute, UK, have already made these nanobodies accessible to researchers at the University of Oxford, UK. These will also soon be widely available to research groups across the world.
Nanobodies are antibodies found in camelids, which are much smaller than human antibodies. According to the researchers, their high stability, small structure and specificity makes them ideal for the purification and stabilisation of proteins and protein structures, prior to imaging.
The team at The Franklin are targeting their work at the S protein, which sits on the outside of the SARS-CoV-2 viral particle and which binds to human cells during an infection. This protein has a specific area – the receptor binding domain (RBD) – that is responsible for this binding action. This makes it an attractive target for future vaccines and therapies.
Nanobodies can stabilise the ‘spike’ to enable better imaging at the atomic scale, using advanced imaging techniques including cryo-electron microscopy (cryo-EM). The nanobodies also allow the RBD to be stabilised when bound to its target, helping researchers better understand how it behaves in the body and how it might interact with new drugs.
The team are also investigating whether the nanobodies they identify, or therapies derived from them, could be used to create highly specific inhibitors, which could contribute to treatments for COVID-19 by preventing the SARS-CoV-2 virus from binding to human cells and causing infection.
Professor James Naismith, Director of the Rosalind Franklin Institute explained: “One single protein often has multiple target sites (epitopes) for the human immune system to produce antibodies to bind to. When antibodies bind to certain epitopes they ‘neutralise’ the virus. This prevents further infection, stopping the virus in its tracks.”
Identifying which nanobodies have ‘binding’ and which have ‘binding and neutralising’ actions is a key next step for the group and one which will see them search for a wider range of nanobodies. The researchers also plan to compare the actions of the new nanobodies to human antibodies derived from patient samples.
Professor Ray Owens, who leads Protein Production UK for The Franklin, said: “There is an unprecedented level of team-work and collaboration globally to image, understand and treat COVID-19. We are working with colleagues at the University of Oxford to use the nanobodies developed here at The Franklin, to gain insights into the structure of the virus that causes COVID -19.”
Related topics
Disease Research, Drug Targets, Imaging, Microscopy, Protein
Related conditions
Coronavirus, Covid-19
Related organisations
Oxford University, Protein Production UK, The Rosalind Franklin Institute
Related people
Professor James Naismith, Professor Ray Owens





good morning
could I use one photo of your site to publish something in Greek language