SARS-CoV-2 model shows Spike protein co-operation
Posted: 1 March 2021 | Victoria Rees (Drug Target Review) | No comments yet
A new coarse-grained model of the complete SARS-CoV-2 virion has revealed potential new ways to combat the coronavirus.
A new multiscale coarse-grained model of the complete SARS-CoV-2 virion, its core genetic material and virion shell, has been developed for the first time using supercomputers. According to its developers, the model offers scientists potential new ways to exploit the virus’s vulnerabilities. The study was conducted at the University of Texas at Austin, US.
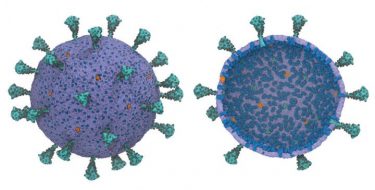
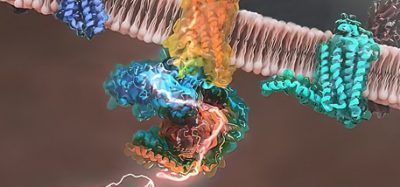
A multiscale model of the complete SARS-CoV-2 virion has been developed for the first time using supercomputers. The model offers scientists the potential for new ways to exploit the virus’s vulnerabilities. Exterior (L) and interior (R) views show the spike protein trimers (teal), glycosylation sites (black), membrane proteins (blue), and pentameric envelope ion channels (orange) [credit: Gregory Voth, University of Chicago].
“We wanted to understand how SARS-CoV-2 works holistically as a whole particle,” said Professor Gregory Voth, corresponding author of the study at the University of Chicago, US. “We developed a bottom-up coarse-grained model where we took information from atomistic-level molecular dynamics simulations and from experiments.”
Voth explained that a coarse-grained model resolves only groups of atoms, versus all-atom simulations, where every single atomic interaction is resolved.
According to the researchers, the early results of the study show how the Spike (S) proteins on the surface of the virus move co-operatively. The S proteins are coupled to each other; when one protein moves another one also moves in response. This motion of the S proteins is informative of how the coronavirus explores and detects the angiotensin-converting enzyme 2 (ACE2) receptors of a potential host cell.
“The paper we published shows the beginnings of how the modes of motion in the S proteins are correlated,” Voth said. “The ultimate goal of the model would be, as a first step, to study the initial virion attractions and interactions with ACE2 receptors on cells and to understand the origins of that attraction and how those proteins work together to go on to the virus fusion process.”
The holistic model started with atomic models of the four main structural elements of the SARS-CoV-2 virion: the S, membrane, nucleocapsid and envelope proteins. These atomic models were then simulated and simplified to generate the complete course-grained model.
Coarse-grained molecular dynamics simulation of the SARS-CoV-2 virion using LAMMPS for 10 × 106 CG time steps [credit: Gregory Voth, University of Chicago].
“What he saw very clearly was the beginning of the dissociation of the S1 subunit of the spike. The whole top part of the spike peels off during fusion,” said study co-author Professor Rommie Amaro, at the University of California, San Diego, US.
One of the first steps of viral fusion with the host cell is this dissociation, where it binds to the ACE2 receptor of the host cell.
“The larger S1 opening movements that they saw with this coarse-grained model was something we had not seen yet in the all-atom molecular dynamics and in fact it would be very difficult for us to see,” Amaro said. “It is a critical part of the function of this protein and the infection process with the host cell. That was an interesting finding.”
“One thing that we are concerned about right now are the UK and the South African SARS-CoV-2 variants. Presumably, with a computational platform like we have developed here, we can rapidly assess those variances, which are changes of the amino acids. We can hopefully rather quickly understand the changes these mutations cause to the virus and then hopefully help in the design of new modified vaccines going forward,” said Voth.
The results of the study were published in the Biophysical Journal.
Related topics
Bioinformatics, Drug Targets, Enzymes, Imaging, Informatics, Protein, Proteomics, Structural Biology
Related conditions
Covid-19
Related organisations
University of California San Diego, University of Chicago, University of Texas at Austin
Related people
Professor Gregory Voth, Professor Rommie Amaro