Drug target to combat three coronavirus diseases identified by researchers
Posted: 16 September 2020 | Victoria Rees (Drug Target Review) | No comments yet
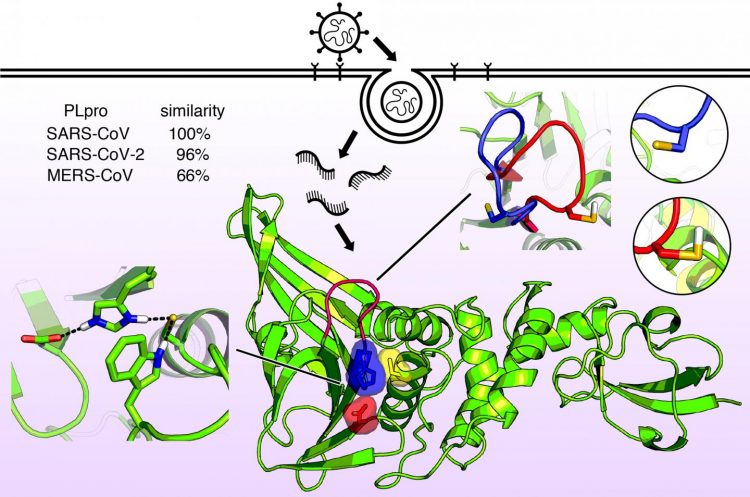
Researchers studying the PLpro binding site of three coronaviruses, including SARS-CoV-2, say the BL2 loop could be targeted by antiviral drugs.

Researchers studying SARS-CoV-2, which is similar in structure to SARS-CoV and MERS-CoV, two other coronaviruses, say they have found a possible pathway to new antiviral drugs to fight all three diseases. The viruses cause, respectively, COVID-19, severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS).
From the University of Maryland School of Pharmacy, US, the researchers conducted molecular-level investigations of these three viruses.
The investigators looked at an enzyme known as the papainlike protease (PLPro), a viral protein that plays a key role in the ability of the coronavirus to replicate itself once inside the body. This protein also plays a role in defeating the host’s immune system, so it provides a particularly attractive target for potential drug treatments.
The protein is nearly identical in SARS-CoV-2 and SARS-CoV but is slightly different in MERS-CoV. Very recently, the first structural X-ray of this enzyme revealed a shape in the catalytic domain somewhat like a hand with a “thumb”, “palm” and “fingers”.
According to the researchers, the thumb and palm come together to form a binding site, where a drug molecule could potentially be captured. The fingers fold down over this region and provide structural integrity that is essential for PLPro activity.

Molecular structure of the papainlike enzyme known as PLPro. This enzyme allows the viruses that cause SARS, MERS, and COVID-19 to infect cells and replicate and suppress the host’s immune function. Now that the enzyme structure is known in detail, new antiviral drugs can be designed [credit: Jack Henderson, University of Maryland School of Pharmacy].
The team discovered small shifts in pH could change the shape of this enzyme through a process known as protonation, where hydrogen ions bind to certain amino acid units in the protein. The researchers say that the protonation state switch is an important energy transduction mechanism.
Co-author Jack Henderson said: “The coronavirus Spike (S) protein, for example, makes use of protonation state switches to induce large conformational changes required for membrane fusion.”
The team say another key feature of the PLpro binding site is a string of amino acid units called the BL2 loop. The investigators found this loop can open or close in SARS coronaviruses when a particular amino acid on the loop is either protonated or deprotonated. In the MERS virus, however, the loop is flexible even without such an amino acid. They say this feature suggests a potential coronavirus drug could target the BL2 loop, causing it to close and tightly bind to a viral inhibitor.
“Our work provides a starting point for further mechanistic investigations using higher-level approaches,” said author Jana Shen.
The results can be found in the Journal of Chemical Physics.
Related topics
Drug Targets, Protein, Proteomics, Research & Development, Target Molecule, Targets
Related conditions
Covid-19, Middle East Respiratory Syndrome (MERS), Severe Acute Respiratory Syndrome (SARS)
Related organisations
Maryland University
Related people
Jack Henderson, Jana Shen