Cell survival mechanisms identified through RNA regulation in central nervous system
Posted: 25 March 2022 | Ria Kakkad (Drug Target Review) | No comments yet
A new study from Niigata University has found a binding protein that is vital for proper development of the central nervous system.
The central nervous system (CNS) is made of the proper proliferation of neural progenitor cells and their differentiation into neurons and glial cells. The transcription factor Olig2 is expressed in neural progenitor cells and is essential for the development of motor neurons and oligodendrocytes and contributes to the proliferation of neural progenitor cells. However, it has remained unknown how Olig2 regulates these diverse biological processes. Scientists from Niigata University, Japan have searched for a novel Olig2 binding factors to explain the developmental mechanisms involved in these factors and Olig2.
In their recent research, published in Cell Death and Differentiation, the researchers identified a new Olig2 binding protein called Ddx20, also known as Genim3 or DP103, that interacts with Olig2 during neural development. Ddx20 is known as a multifunctional molecule that regulates transcription, RNA splicing and protein translation. Furthermore, Ddx20 interacts with SMN, a causative gene of spinal muscular atrophy (SMA) and is deeply involved in the pathogenesis of SMA.
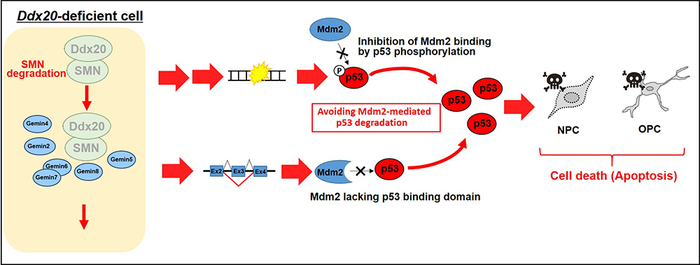
In Ddx20-deficient cells, the SMN complex collapses due to destabilisation of SMN protein, resulting in abnormal RNA splicing process (pink background). This leads to DNA damage and abnormal splicing of Mdm2 mRNA and activation of the p53 pathway due to reduced p53 proteolysis, resulting in apoptosis (cell death) of neural progenitor cells and oligodendrocyte progenitor cells.
[Credit: Niigata University]
The study involved generating Ddx20 conditional knockout mice to investigate the function of Ddx20 during CNS development. They found that apoptosis occurs rapidly in neural progenitor cells and oligodendrocyte progenitor cells. The mechanism responsible for the cell death was investigated and excessive nuclear accumulation of p53, a tumour suppressor gene product, was found. The researchers also found that interestingly, in Ddx20-deficient mice, the SMN protein was destabilised and the RNA splicing mechanisms were abnormal, leading to splicing dysregulation of Mdm2, a p53 inhibitor.
They proceeded to further understand the mechanism by which Olig2 contributes to neural progenitor cell proliferation and survival. In these experiments, the scientists found that Ddx20 levels and SMN protein levels were lower in Olig2-deficient mice than in wild-type mice. In addition, some spliceosomal RNA and Mdm2 splicing were dysregulated in Olig2-deficient mice than in wild-type mice. Lastly, the p53 was more activated in Olig2-deficient mice than in wild-type mice and the team found that Olig2 maintains the Ddx20-SMN complex involved in the regulation of Mdm2 splicing, which in turn suppress p53 activation.
“This study highlights that a transcriptional factor, Olig2, affects not only transcriptional regulation but also RNA metabolism through Ddx20 stabilisation, revealing the diverse functions of Olig2. Importantly, since Olig2 has been reported to be involved not only in neural development but also in the progression of glioma and melanoma. On the other hand, Ddx20 has also been known as an initiator of various cancers. Therefore, the interaction of these two factors may play a key role in the development and progression of the cancers. We believe that further research might provide clues to elucidate the etiology of congenital neurological diseases and cancers, including the development of therapeutic strategies against them,” concluded Professor Hirohide Takebayashi, who led this research.
Related topics
Cell-based assays, Central Nervous System (CNS), Disease Research, Drug Leads, Molecular Biology, Molecular Modelling, Molecular Targets, Neurons, Protein, Research & Development, RNAs
Related conditions
congenital neurological diseases
Related organisations
Niigata University
Related people
Professor Hirohide Takebayashi